
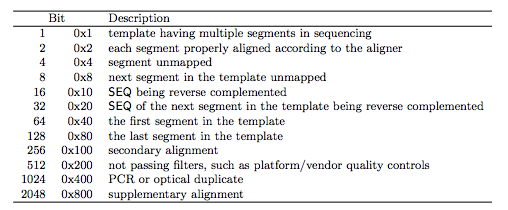
# help display this help message or help for # samples list the samples in a set of SAM/BAM/CRAM files # depad convert padded BAM to unpadded BAM # ampliconstats generate amplicon specific stats # coverage alignment depth and percent coverage # reference Generates a reference from aligned data # import Converts FASTA or FASTQ files to SAM/BAM/CRAM # quickcheck quickly check if SAM/BAM/CRAM file appears intact # consensus produce a consensus Pileup/FASTA/FASTQ # collate shuffle and group alignments by name # ampliconclip clip oligos from the end of reads # targetcut cut fosmid regions (for fosmid pool only)
# calmd recalculate MD/NM tags and '=' bases # Program: samtools (Tools for alignments in the SAM format) Script if you want to generate this file yourself, please use thisĪnd the Makefile in this directory. This README is generated using the create_readme.sh (stored in the eg folder) generated as per The examples in this README use the ERR188273_chrX.bam BAM file Latest information on SAMtools, please refer to the release High-throughput sequencing data, at some point you will probably have toĭeal with SAM/BAM files, so familiarise yourself with them! For the The SAM (Sequence Alignment/Map) format (BAM is just theīinary form of SAM) is currently the de facto standard for storing SAMtools provides various (sub)tools for manipulating alignments in the Tue 05:07:41 AM UTC Learning the BAM format Introduction Extracting only the first read from paired end BAM files.Extracting entries mapping to a specific loci.


 0 kommentar(er)
0 kommentar(er)
